Gene Essentiality Prediction Using Topological Features From Metabolic Networks
source:https://ieeexplore.ieee.org/document/8575595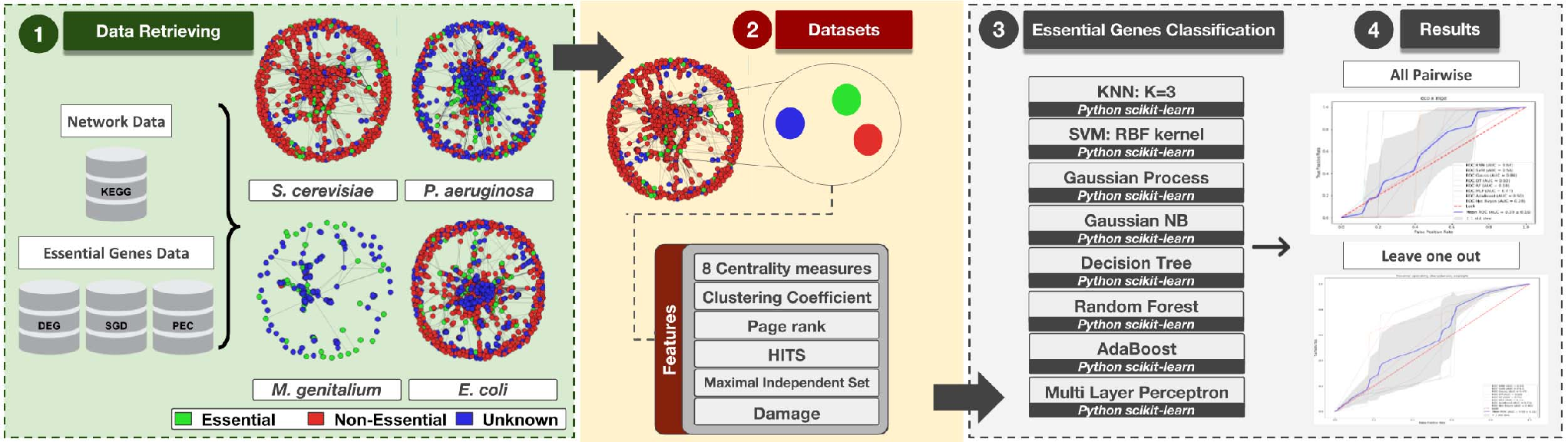
source:https://ieeexplore.ieee.org/document/8575595
source:https://www.sciencedirect.com/science/article/pii/S1934590920305427?via%3Dihub#undfig1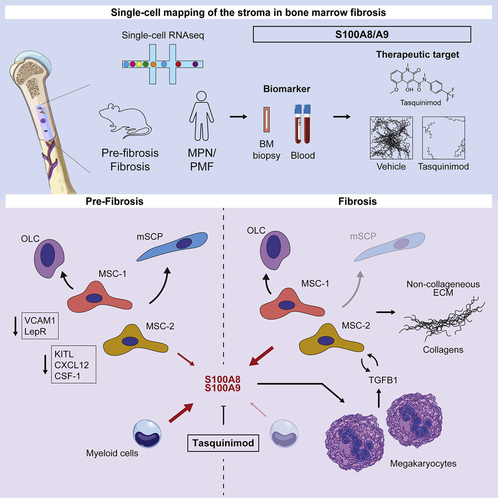
source:https://www.biorxiv.org/content/10.1101/2021.01.20.427390v2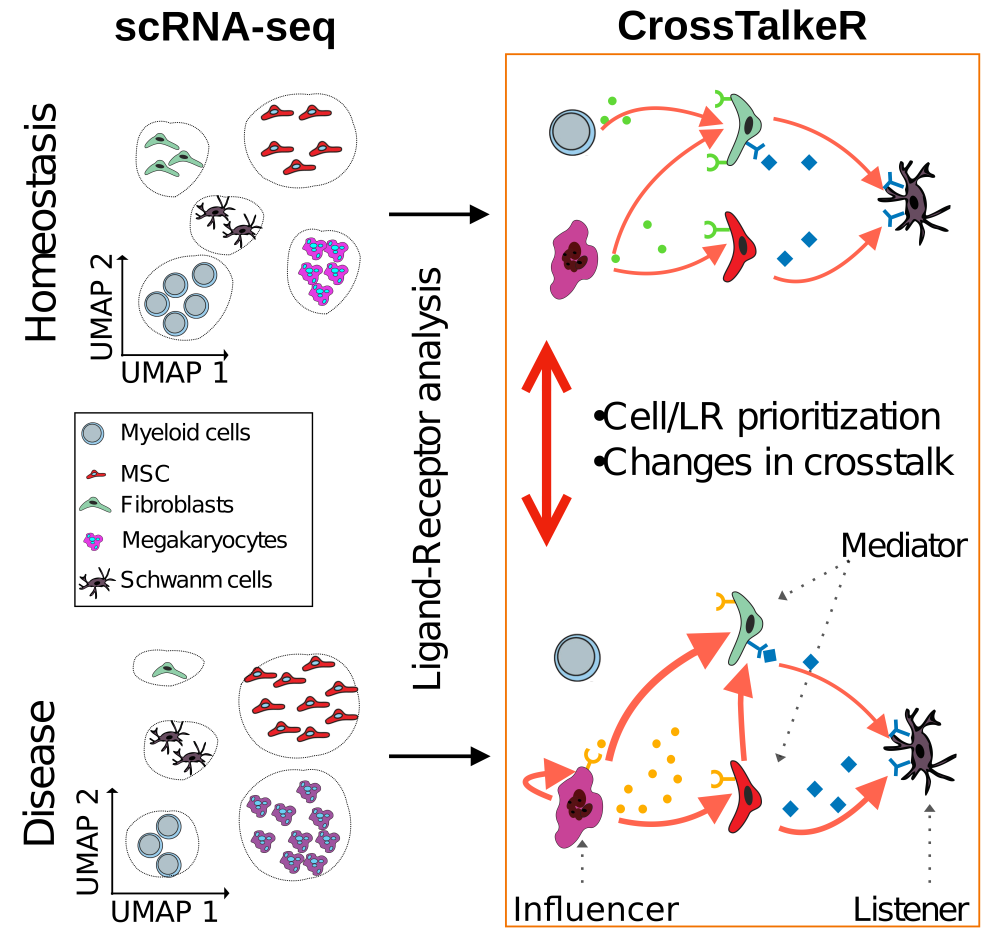
Published in Encontro Nacional de Engenharia e Desenvolvimento Social, XIV Encontro Nacional de Engenharia e Desenvolvimento Social, 2017
Recommended citation: Aono, A.H., Nagai, J.S., Santos, S.R.S., Cespedes, J.G. and da Silva, L.F., 2017. PluviApp: Ferramenta Web para análise e visualização de dados pluviométricos. Anais dos Encontros Nacionais de Engenharia e Desenvolvimento Social-ISSN 2594-7060, 14(1). http://www.eneds.net/anais/index.php/edicoes/eneds2017/paper/view/543/438
Published in KDMiLe – Proceedings Applications Track, 2018
Recommended citation: Aono, A.H., de Oliveira, R.M., Franchi, B.O., Nagai, J.S., Paz, H.E.S., Chaves, A.A. and Martins, C.B., 2018. A Biased Random-key Genetic Algorithm with Local Search Applied to Unsupervised Clustering of Cultural Data Applications Track. https://www.researchgate.net/profile/Alexandre-Aono/publication/330325486_A_Biased_Random-key_Genetic_Algorithm_with_Local_Search_Applied_to_Unsupervised_Clustering_of_Cultural_Data/links/5c39205692851c22a36e4535/A-Biased-Random-key-Genetic-Algorithm-with-Local-Search-Applied-to-Unsupervised-Clustering-of-Cultural-Data.pdf
Published in 2018 7th Brazilian Conference on Intelligent Systems (BRACIS), 2018
Recommended citation: Nagai, J.S., Sousa, H., Aono, A.H., Lorena, A.C. and Kuroshu, R.M., 2018, October. Gene essentiality prediction using topological features from metabolic networks. In 2018 7th Brazilian Conference on Intelligent Systems (BRACIS) (pp. 91-96). IEEE. https://ieeexplore.ieee.org/abstract/document/8575595
Published in biorxiv, 2019
Recommended citation: Aono, A.H., Nagai, J.S., da SM Dickel, G., Marinho, R.C., de Oliveira, P.E. and Faria, F.A., 2019. A stomata classification and detection system in microscope images of maize cultivars. bioRxiv, p.538165. https://www.biorxiv.org/content/biorxiv/early/2019/02/01/538165.full.pdf
Published in Scientific Reports, 2020
Recommended citation: Aono, A.H., Costa, E.A., Rody, H.V.S., Nagai, J.S., Pimenta, R.J.G., Mancini, M.C., dos Santos, F.R.C., Pinto, L.R., de Andrade Landell, M.G., de Souza, A.P. and Kuroshu, R.M., 2020. Machine learning approaches reveal genomic regions associated with sugarcane brown rust resistance.Sci Rep 10, 20057 (2020). https://www.nature.com/articles/s41598-020-77063-5
Published in Cell Stem Cell, 2020
Recommended citation: Leimkühler, N.B., Gleitz, H.F., Ronghui, L., Snoeren, I.A., Fuchs, S.N., Nagai, J.S., Banjanin, B., Lam, K.H., Vogl, T., Kuppe, C. and Stalmann, U.S., 2020. Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell. https://www.sciencedirect.com/science/article/pii/S1934590920305427
Published in FrontiersinImmunology, 2021
Recommended citation: Schreibing, Felix, Monica Hannani, Fabio Ticconi, Eleanor Fewings, James Shiniti Nagai, Matthias Begemann, Christoph Kuppe et al. "Dissecting CD8+ T cell pathology of severe SARS-CoV-2 infection by single-cell epitope mapping." bioRxiv (2021). https://www.frontiersin.org/articles/10.3389/fimmu.2022.1066176/full
Published in Frontiers in Plant Science, 2021
Recommended citation: Aono, Alexandre Hild, Ricardo José Gonzaga Pimenta, Ana Letycia Basso Garcia, Fernando Henrique Correr, Guilherme Kenichi Hosaka, Marishani Marin Carrasco, Claudio Benicio Cardoso-Silva et al. "The Wild Sugarcane and Sorghum Kinomes: Insights into Expansion, Diversification and Expression Patterns." Frontiers in Plant Science 12 (2021): 589. https://www.frontiersin.org/articles/10.3389/fpls.2021.668623/abstract
Published in Bioinformatics, 2021
Recommended citation: Nagai, James Shiniti, Nils Leimkuehler, Michael Schaub, Rebekka Schneider, and Ivan Gesteira Costa Filho. "CrossTalkeR: Analysis and Visualisation of Ligand Receptor Networks." Bioinformatics (2021). https://doi.org/10.1093/bioinformatics/btab370,https://www.biorxiv.org/content/10.1101/2021.01.20.427390v1
Published in Science of The Total Enviroment, 2021
Recommended citation: Ishimoto, Caroline Kie, Alexandre Hild Aono, James Shiniti Nagai, Hério Sousa, Ana Roberta Lima Miranda, Vania Maria Maciel Melo, Lucas William Mendes et al. "Microbial co-occurrence network and its key microorganisms in soil with permanent application of composted tannery sludge." Science of The Total Environment (2021): 147945. https://www.sciencedirect.com/science/article/abs/pii/S0048969721030163
Published in Nature Communication, 2022
Recommended citation: Dimitrov, D., Türei, D., Garrido-Rodriguez, M., Burmedi, P. L., Nagai, J. S., Boys, C., ... & Saez-Rodriguez, J. (2022). Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nature Communications, 13(1), 1-13. https://www.nature.com/articles/s41467-022-30755-0
Published in Cell Stem Cell, 2022
Recommended citation: Jansen, J., Reimer, K. C., Nagai, J. S., Varghese, F. S., Overheul, G. J., de Beer, M., ... & Powell, A. (2022). SARS-CoV-2 infects the human kidney and drives fibrosis in kidney organoids. Cell stem cell, 29(2), 217-231. https://www.sciencedirect.com/science/article/pii/S1934590921005208
Published in Development, 2022
Recommended citation: Jansen, J., van den Berge, B. T., van den Broek, M., Maas, R. J., Daviran, D., Willemsen, B.,..., Nagai. J. S.,... & Smeets, B. (2022). Human pluripotent stem cell-derived kidney organoids for personalized congenital and idiopathic nephrotic syndrome modeling. Development, 149(9), dev200198. https://journals.biologists.com/dev/article/149/9/dev200198/275303/Human-pluripotent-stem-cell-derived-kidney
Published in Experimental Hematology, 2022
Recommended citation: Stalmann, U. S., Banjanin, B., Snoeren, I. A., Nagai, J. S., Leimkühler, N. B., Li, R., ... & Schneider, R. K. (2022). Single-cell analysis of cultured bone marrow stromal cells reveals high similarity to fibroblasts in situ. Experimental Hematology, 110, 28-33. https://www.sciencedirect.com/science/article/pii/S0301472X22001308
Published in Nature Communication, 2022
Recommended citation: Peisker, F., Halder, M., Nagai, J., Ziegler, S., Kaesler, N., Hoeft, K., ... & Kramann, R. (2022). Mapping the cardiac vascular niche in heart failure. Nature Communications, 13(1), 1-20. https://www.nature.com/articles/s41467-022-30682-0
Published in Nature, 2022
Recommended citation: Kuppe, C., Ramirez Flores, R. O., Li, Z., Hayat, S., Levinson, R. T., Liao, X., ... & Kramann, R. (2022). Spatial multi-omic map of human myocardial infarction. Nature, 1-12. https://www.nature.com/articles/s41586-022-05060-x
Published in Nature Genetics, 2022
Recommended citation: Xu, Y., Kuppe, C., Perales-Patón, J., Hayat, S., Kranz, J., Abdallah, A. T., ... & Kramann, R. (2022). Adult human kidney organoids originate from CD24+ cells and represent an advanced model for adult polycystic kidney disease. Nature genetics, 54(11), 1690-1701. https://www.nature.com/articles/s41588-022-01202-z
Published in biorxiv, 2022
Recommended citation: Joodaki, M., Shaigan, M., Parra, V., Buelow, R. D., Kuppe, C., Hölscher, D. L., ... & Costa, I. G. (2022). Detection of PatIent-Level distances from single cell genomics and pathomics data with Optimal Transport (PILOT). bioRxiv, 2022-12. https://www.cell.com/cell-reports/pdf/S2211-1247(23)00142-0.pdf
Published in BioinformaticsAdvances, 2023
Recommended citation: Li, Z., Nagai, J. S., Kuppe, C., Kramann, R., & Costa, I. G. (2023). scMEGA: single-cell multi-omic enhancer-based gene regulatory network inference. Bioinformatics Advances, 3(1), vbad003. https://academic.oup.com/bioinformaticsadvances/article/3/1/vbad003/6986159
Published in CellReports, 2023
Recommended citation: Hoeft, K., Schaefer, G. J., Kim, H., Schumacher, D., Bleckwehl, T., Long, Q., ... & Kramann, R. (2023). Platelet-instructed SPP1+ macrophages drive myofibroblast activation in fibrosis in a CXCL4-dependent manner. Cell Reports, 42(2). https://www.cell.com/cell-reports/pdf/S2211-1247(23)00142-0.pdf
Published:
CostaLab group meeting presentation
Published:
CrossTalkeR is a framework for network analysis and visualisation of LR networks. CrossTalkeR identifies relevant ligands, receptors and cell types contributing to changes in cell communication when contrasting two biological states: disease vs. homeostasis. A case study on scRNA-seq of human myeloproliferative neoplasms reinforces the strengths of CrossTalkeR for characterisation of changes in cellular crosstalk in disease state.
Published:
scMEGA Fantastic work from Zhijian Li(https://lzj1769.github.io/). scMEGA(Single-cell Multiomic Enhancer-based Gene regulAtory network inference) is a R package designed to infer gene regulatory network by using single cell multi-omics data. It is based on several popular packages for single cell RNA/ATAC-seq data analysis, particularly, Seurat, Signac, and ArchR. Picture below showed the principal of scMEGA for linking genes to TFs and building a gene regualtory network.